Christiaen Lab Research
Overview
During animal development, tissues and organs assume shapes and positions that are largely determined by the species-specific genetic blueprint. These macroscopic events are powered by a variety of cells that divide, change shape and move in response to cell-cell communications and lineage-specific genetic programs. Our overarching goal is to reach a system’s level understanding of how tissue-specific regulatory programs and cell-cell communications coordinate the fate choices and cellular behaviors that underlie animal development, regeneration and evolution.
We focus on cardiopharyngeal lineages, which produce both cardiomyocytes of the second heart field and head muscles from Mesp1+ anterior mesoderm progenitors. The cardiopharyngeal paradigm casts new light on certain congenital diseases characterized by the co-occurence of cardiac and craniofacial defects, such as the DiGeorge/22q11.2 deletion syndrome. The latter is thought to emerge from haploinsufficiency of TBX1, a key determinant of cardiopharyngeal progenitors. This illustrates the biomedical relevance of our work on the regulation of cardiopharyngeal multipotency and early heart vs. pharyngeal muscle fate choices. Circumventing the complexity of vertebrates, we use embryos and larvae of the tunicate Ciona to study cardiopharyngeal development with high spatial and temporal resolution. In ascidians like Ciona, every invariant division and migratory event is mapped onto a stereotyped and evolutionarily conserved developmental sequence. We strive to develop state-of-the-art methods to study Ciona development, including targeted genome engineering with CRISPR/Cas9, single cell genomics, quantitative imaging and mathematical modeling. Leveraging the simplicity of the Ciona model and our extensive experimental toolkit, we pursue a system’s level understanding of cellular behaviors with a focus on collective polarity, directed migration, and oriented and asymmetric cell divisions.
Finally, we are expanding the scope of our research towards (1) the evolution of cardiopharyngeal development through comparative studies in tunicates and vertebrates, including mammalian stem cell models; (2) the regenerative potential of cardiopharyngeal structures in tunicates and their relationships with other endomesodermal systems; and (3) environmentally-relevant problems in developmental systems biology.
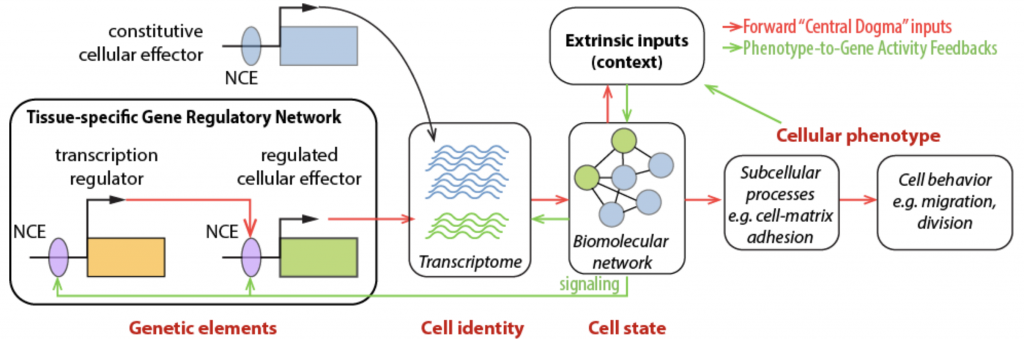
Fig.1.
An extended central dogma for developmental cell behavior.
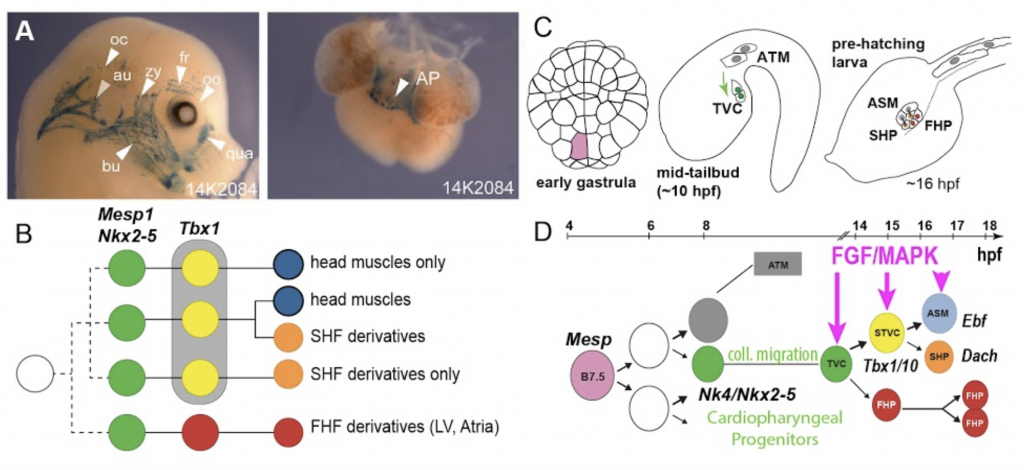
Fig.2.
The cardiopharyngeal mesoderm.
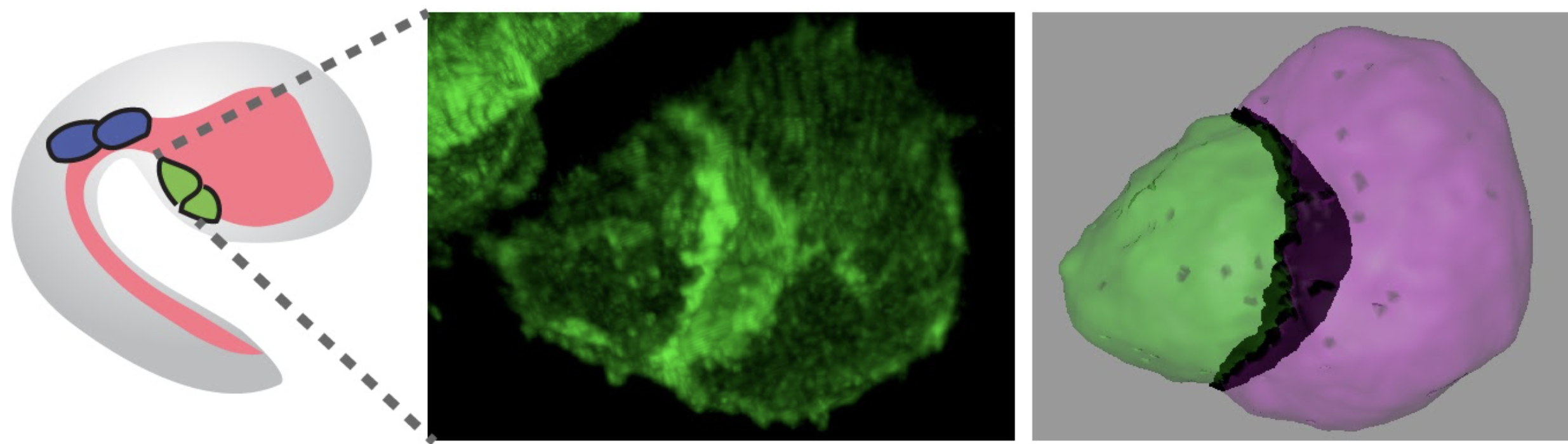
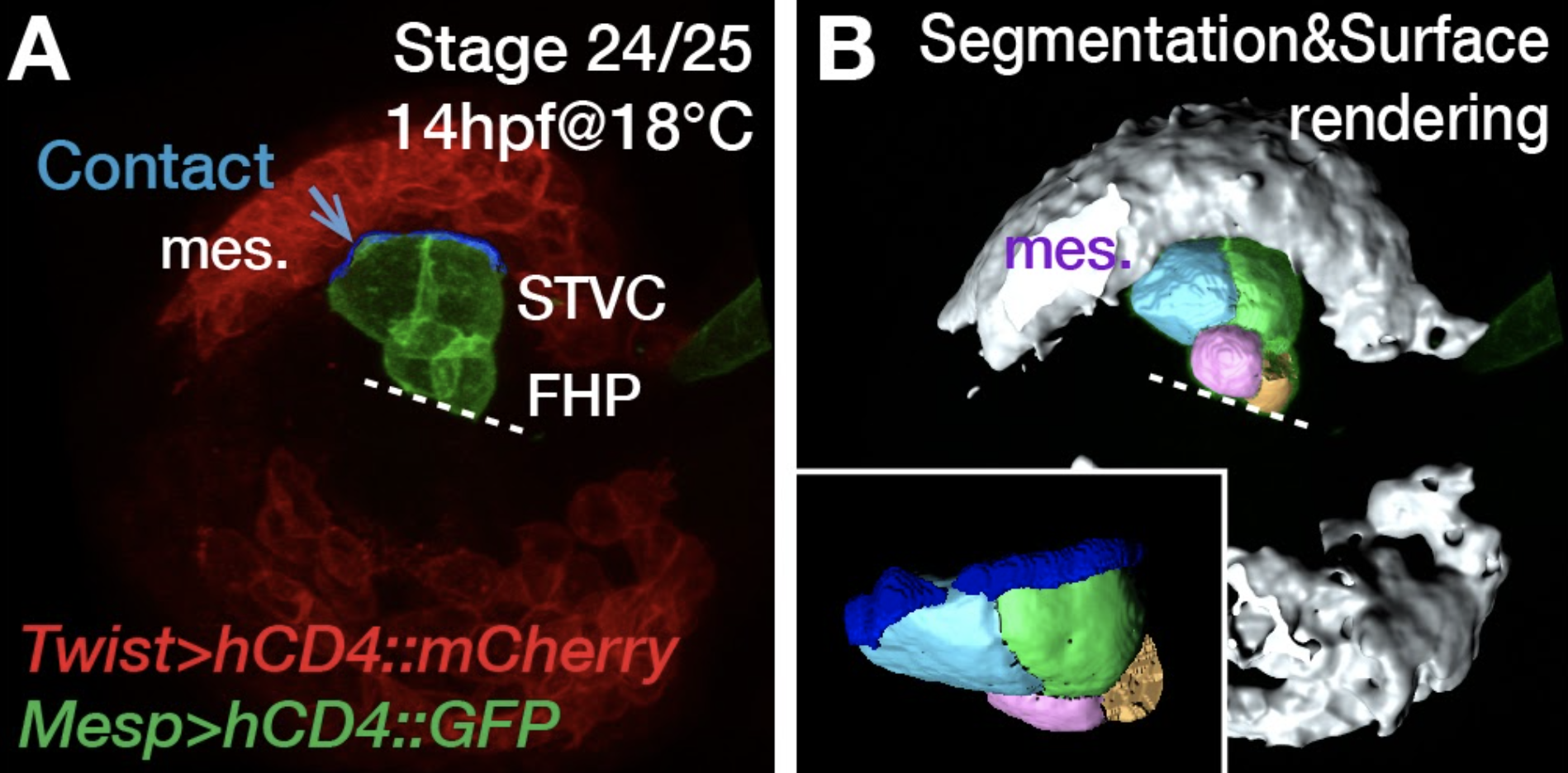
Fig.3.
Multipotent cardiopharyngeal progenitors offer the simplest possible model for collective cell migrations.
Christiaen Lab research
Systems biology of cellular behaviors
Pursuing our overarching goal to connect gene regulatory networks that control cell fates with the cellular behaviors underlying morphogenesis, we first sought to combine fluorescence activated cell sorting (FACS) and transcriptome profiling to identify genes upregulated in migratory cardiopharyngeal progenitors, following induction by FGF-MAPK signaling and the onset of Foxf expression1–3. These progenitors, also called trunk ventral cells (TVCs) in ascidians, represent the simplest possible collective cell migration with only two cells on either side of the embryo, which nonetheless polarize collectively and migrate with defined leader and trailer cells4,5.
We developed methods for targeted perturbations, live imaging, and quantitative analyses, which allowed us to begin to characterize the surrounding tissues that influence cardiopharyngeal progenitor behavior4, as well as the impact of regulated intrinsic cellular effector genes on collective polarity and directed migration5.
We proposed a conceptual framework for the regulation of developmental cell behavior, which postulates that factors modulating the interpretation and response to extrinsic cues are privileged targets for the transcriptional control of tissue-specific cellular behaviors6. We reasoned that cellular effectors function dynamically and in subcellular pools that are better understood in live cells using biosensors and optogenetic tools, to achieve the most appropriate spatial and temporal resolution.
Focusing on collective polarity, directed migration, and asymmetric and oriented cell divisions, we pursue the following research projects:
- Mathematical modeling of forces and gene activities underlying collective migration and interactions with surrounding tissues. (Y. Bernadskaya, collaboration with A. Mogilner)
- CRISPR/Cas9 screening, High-content image analysis, and biomolecular network modeling. (M. Failla & K. Wiechecki)
- Biosensors and optogenetics to study the subcellular modules underlying developmental cell behaviors.
Christiaen Lab research
The regulation of multipotency and cell fate choices
In 2010, we first reported that multipotent cardiopharyngeal progenitors produce first and second heart lineages, and pharyngeal muscle precursors following simple and stereotyped divisions patterns7. Since then, we have sought to characterize the general principles, and cellular and molecular underpinnings of cardiopharyngeal multipotency and early fate choices.
Using whole mount fluorescent in situ hybridization, targeted perturbations, including CRISPR-Cas9-mediated mutagenesis, and single cell genomics, we found that transcriptional multilineage priming and regulatory cross-antagonisms between cardiac and pharyngeal muscle programs are hallmarks of early cardiopharyngeal development, and involve such conserved regulators as Nk4/Nkx2-5, Tbx1/10 and Ebf (aka. Collier/Olf/Ebf or COE)8–10. This multilineage priming extends to widespread and cardiopharyngeal-specific chromatin accessibility, which is largely governed by FGF-MAPK signaling and one of its downstream targets, the transcription factor Foxf11.
Asymmetric maintenance of FGF-MAPK signaling and feedforward circuits are coupled with oriented and unequal cell cleavages to control progressive and differential fate choices10,12. Finally, we reasoned that extensive multilineage priming demands widespread post-transcriptional regulatory mechanisms to ensure rapid transcriptome reprogramming and fate choices.
We are currently pursuing the following outstanding questions regarding cardiopharyngeal multipotency and early fate choices:
- A cardiopharyngeal niche, which provides symmetry breaking cues for asymmetric signaling. (N. Kaplan)
- Coupling cell cycle progression, de novo gene expression and fate decisions.
- Termination of FGF-MAPK signaling and commitment to a cardiac identity.
- Maturation of multipotent progenitors and the acquisition of cardiopharyngeal competence. (W. Wang & A. Tjärnberg)
- Gene regulatory network models) (A.Tjärnberg, collaboration with R. Bonneau). (JOB)
- Post-transcriptional regulatory mechanisms and proteomics. (B. Vitrinel, collaboration with C. Vogel)
Fig.4.
The cardiopharyngeal niche.
Christiaen Lab research
Pharyngeal muscle morphogenesis and differentiation, cardiac organogenesis and regeneration
Following early commitment to either a pharyngeal muscle or cardiac identity, cells undergo tissue-specific morphogenesis and differentiation.
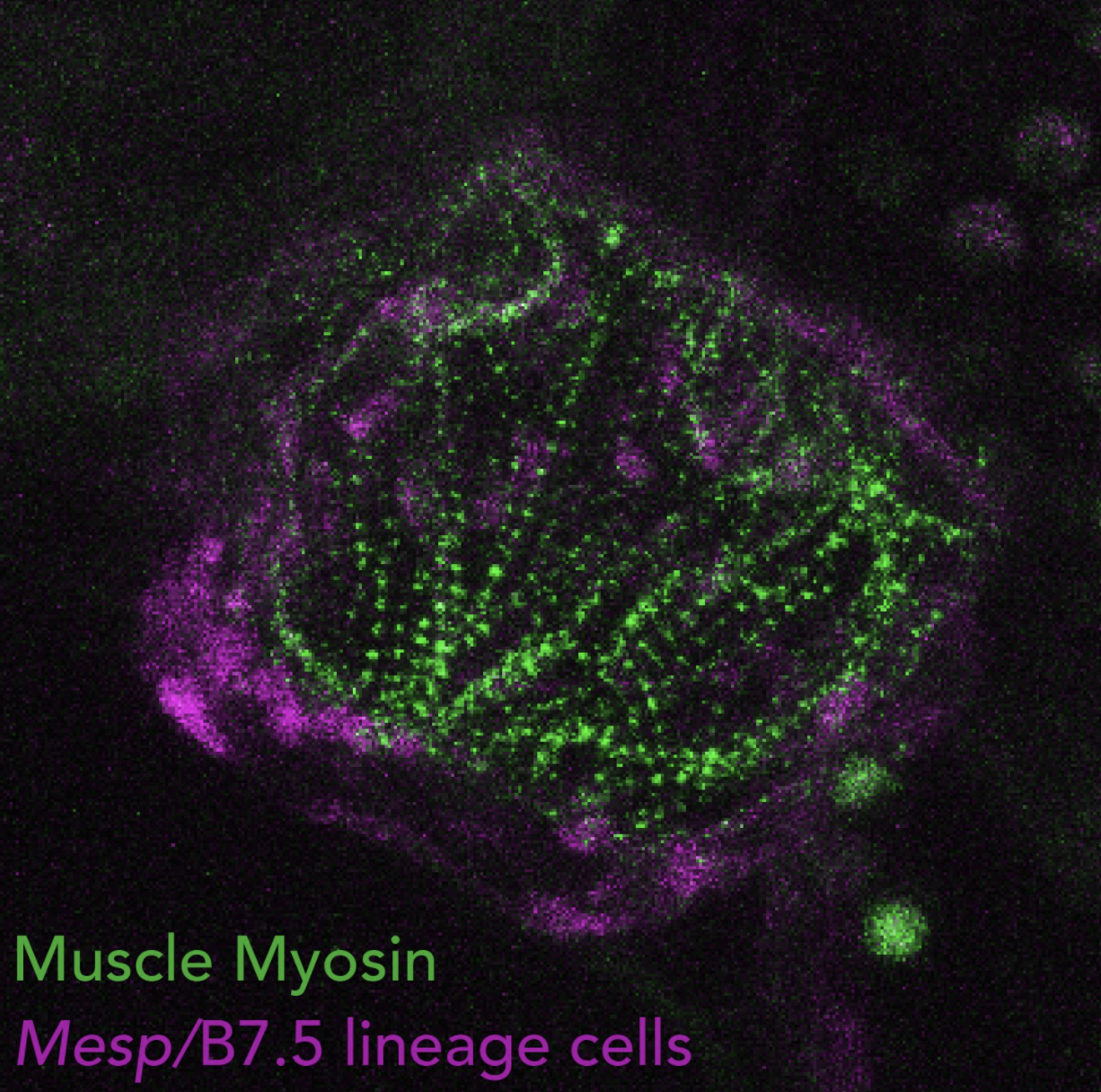
Four pharyngeal muscle precursors, aka Atrial Siphon Muscle Precursors (ASMPs) migrate collectively from the ventral side of the swimming larva to the dorsal atrial siphon placode, a presumed homolog of the vertebrate otic placode, and occasionally divide to form a conspicuous ring of 4 to 8 cells7,8.
On either side of the larva, the 4 ASMPs are born from 2 Atrial Siphon Muscle Founder cells (ASMFs) and align along the anterior-posterior axis, where pairs of siblings each form one inner ASMP (iASMP) and one outer ASMP (oASMP). Remarkably, oASMPs maintain detectable levels of Mrf mRNA, the Ciona homolog of Myod, divide only once and are the first muscle cells to elongate and differentiate. By contrast, iASMPs downregulate Mrf expression in response to Notch signaling, and later divide many more times to produce the majority of ASM and longitudinal body wall muscles, in a manner analogous to undifferentiated stem-cell-like muscle progenitors in vertebrates8.
Outstanding questions remain concerning:
- The mechanisms that govern the collective migration of pharyngeal muscle precursor cells and their association with the ectodermal placode.
- The potential of undifferentiated muscle progenitors to produce satellite-like muscle stem cells, and promote muscle regeneration.
Distinct but related first and second cardiac lineages contribute cells to the beating heart8–10. Although the heart differentiates where cells are first born (with some left-right asymmetry13), on the ventral side of the larva, it transitions from a monolayer of 4 x 4 cells to a 2-layer cone-like open compartment, which starts to beat ~72 hours post-fertilization10. Single cell RNA sequencing (scRNA-seq) and lineage-tracing indicated that the first and second lineages produce primarily cardiomyocytes (CM) of the inner layer (closer to the lumen) vs. non-CM cells in the outer layer, respectively10. Since Ciona lacks a bona fide endocardium, the CMs form a myoepithelium that is in direct contact with the blood14. Outstanding questions remain regarding:
- Heart morphogenesis, connection with the vasculature and blood development.
- The regenerative potential of the heart. (K. Schuster)
Christiaen Lab research
Cardiopharyngeal evolution and comparative studies
We coined the term “cardiopharyngeal” in a study that highlighted the evolutionary parallels between the cardiogenic B7.5 lineage of ascidians and the heart lineages of vertebrates, especially the second heart field and its developmental connection to branchiomeric head muscles7. We have since refined and expanded our understanding of the conserved features of cardiopharyngeal development in olfactores, the monophyletic group that combines vertebrates and their closest relatives, the tunicates15.
We also postulated that the cardiopharyngeal mesoderm took part in the evolution of the vertebrate “new head”16. More recently, emerging methods for comparative single cell genomics have allowed us to identify markers of cardiopharyngeal fates conserved between Ciona and the mouse10.
We identified novel markers and determinants of pharyngeal muscles and of the second heart field, the transcription factors Ebf1 and Dach1, respectively, thus illustrating the potential of studies using Ciona to inform cardiopharyngeal developmental biology in mammals.
We pursue comparative studies with vertebrate models in several directions:
- Comparative gene expression between Ciona and the mouse. (collaboration with R. Kelly, and Leducq Network)
- Cross-species enhancer and regulatory gene analysis. (collaborations with C. Mosimann, N. Palpant, I. Scott)
- FUTURE: mammalian gastruloid models of cardiopharyngeal development.
The tunicates form a diverse group of species on its own merit, and with an intriguing evolutionary property: ascidians, including species that evolved separately for more than 350 M years, retained virtually invariant early embryogenesis despite profound divergence in genome sequences.
Extensive developmental systems drift accounts for the apparent discrepancy between divergent genomic sequences and conserved embryonic phenotypes. Specifically, we reported that key cell division, migration and regulatory gene expression in the cardiopharyngeal lineage are conserved between Ciona, a Phlebobranch, and distant relatives among the Stolidobranchs, species of the Molgula genus17.
However, cross-species reporter assays indicated that enhancer structure and function are subjected to developmental systems drift and largely unintelligible between species.
Likewise, we analyzed the A7.6 lineage in Ciona, which produces oral siphon muscles (OSM) almost identical to the B7.5-derived atrial siphon muscles. This indicated that a shared pharyngeal muscle program, defined by the co-expression of Tbx1/10, Ebf and Mrf, is deployed in OSM progenitor cells, but the relative timing and regulatory relationships between key determinants differ between the two lineages18.
Like developmental systems drift, these observations illustrate the plasticity of regulatory programs governing conserved gene expression profiles and cell identities.
Christiaen Lab research
Developmental systems genetics from the lab to the ocean.
Building on the rich genomic resources that propelled the Ciona model into twenty-first century Science19, we continuously strive to adapt state-of-the-art methods for functional genomics and developmental cell and systems biology.
We pioneered the combination of FACS with microarrays1, RNA-seq and scRNA-seq10 for lineage-specific transcriptome profiling, and with ATAC-seq for chromatin accessibility11. We adapted the RNAi5 and CRISPR/Cas9 systems20,21 for lineage-specific loss-of-function assays. We established a 2-component transgenic system based on LexA/LexAop18, which augments the combinatorial potential for genetic manipulations in Ciona.
With the advent of image- and sequence-based methods that empower quantitative assays, the need for correspondingly precise genetic tools is more pressing. These efforts were pioneered for many years by Yasunori Sasakura and colleagues, who established methods for stable transgenesis.
We recently implemented a simple culturing system, in the hope to develop precise genome engineering methods (aka “knock-in”), and cultured hybrids between Ciona intestinalis and Ciona robusta, as a proof of principle22. Ongoing and prospective method development projects include:
- Germline biology and CRISPR/Cas9-mediated knock-ins. (N. Ohta)
- Light and chemical-inducible systems for transgene expression.
Finally, our ability to culture wild animals and inter-specific hybrids opens new opportunities to develop quantitative genetics methods to study complex traits in Ciona and/or other tunicates.
Specifically, we envision to deploy powerful functional genomic methods, including scRNA-seq and CRISPR/Cas9-mediated genome editing, and tap the potential of natural variation in wild populations to study environmentally-relevant problems related to ocean health and the impact of global climate changes on marine invertebrates, like the tunicates.
References
2. Davidson, B., Shi, W., Beh, J., Christiaen, L. & Levine, M. FGF signaling delineates the cardiac progenitor field in the simple chordate, Ciona intestinalis. Genes Dev. 20, 2728–2738 (2006).
3. Beh, J., Shi, W., Levine, M., Davidson, B. & Christiaen, L. FoxF is essential for FGF-induced migration of heart progenitor cells in the ascidian Ciona intestinalis. Development 134, 3297–3305 (2007).
4. Gline, S., Kaplan, N., Bernadskaya, Y., Abdu, Y. & Christiaen, L. Surrounding tissues canalize motile cardiopharyngeal progenitors towards collective polarity and directed migration. Development 142, 544–554 (2015).
5. Bernadskaya, Y. Y., Brahmbhatt, S., Gline, S. E., Wang, W. & Christiaen, L. Discoidin-domain receptor coordinates cell-matrix adhesion and collective polarity in migratory cardiopharyngeal progenitors. Nat. Commun. 10, 57 (2019).
6. Bernadskaya, Y. & Christiaen, L. Transcriptional Control of Developmental Cell Behaviors. Annu. Rev. Cell Dev. Biol. 32, 77–101 (2016).
7. Stolfi, A. et al. Early chordate origins of the vertebrate second heart field. Science 329, 565–568 (2010).
8. Razy-Krajka, F. et al. Collier/OLF/EBF-dependent transcriptional dynamics control pharyngeal muscle specification from primed cardiopharyngeal progenitors. Dev. Cell 29, 263–276 (2014).
9. Wang, W., Razy-Krajka, F., Siu, E., Ketcham, A. & Christiaen, L. NK4 antagonizes Tbx1/10 to promote cardiac versus pharyngeal muscle fate in the ascidian second heart field. PLoS Biol. 11, e1001725 (2013).
10. Wang, W. et al. A single-cell transcriptional roadmap for cardiopharyngeal fate diversification. Nature Cell Biology vol. 21 674–686 (2019).
11. Racioppi, C., Wiechecki, K. A. & Christiaen, L. Combinatorial chromatin dynamics foster accurate cardiopharyngeal fate choices. Elife 8, (2019).
12. Razy-Krajka, F. et al. An FGF-driven feed-forward circuit patterns the cardiopharyngeal mesoderm in space and time. Elife 7, (2018).
13. Palmquist, K. & Davidson, B. Establishment of lateral organ asymmetries in the invertebrate chordate, Ciona intestinalis. Evodevo 8, 12 (2017).
14. Evans Anderson, H. & Christiaen, L. Ciona as a Simple Chordate Model for Heart Development and Regeneration. Journal of Cardiovascular Development and Disease 3, 25 (2016).
15. Kaplan, N., Razy-Krajka, F. & Christiaen, L. Regulation and evolution of cardiopharyngeal cell identity and behavior: insights from simple chordates. Curr. Opin. Genet. Dev. 32, 119–128 (2015).
16. Diogo, R. et al. A new heart for a new head in vertebrate cardiopharyngeal evolution. Nature 520, 466–473 (2015).
17. Stolfi, A. et al. Divergent mechanisms regulate conserved cardiopharyngeal development and gene expression in distantly related ascidians. Elife 3, e03728 (2014).
18. Tolkin, T. & Christiaen, L. Rewiring of an ancestral Tbx1/10-Ebf-Mrf network for pharyngeal muscle specification in distinct embryonic lineages. Development 143, 3852–3862 (2016).
19. Dehal, P. et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science 298, 2157–2167 (2002).
20. Stolfi, A., Gandhi, S., Salek, F. & Christiaen, L. Tissue-specific genome editing in Ciona embryos by CRISPR/Cas9. Development 141, 4115–4120 (2014).
21. Gandhi, S., Haeussler, M., Razy-Krajka, F., Christiaen, L. & Stolfi, A. Evaluation and rational design of guide RNAs for efficient CRISPR/Cas9-mediated mutagenesis in Ciona. Dev. Biol. 425, 8–20 (2017).
22. Ohta, N., Kaplan, N., Ng, J. T., Gravez, B. J. & Christiaen, L. Asymmetric Fitness of Second-Generation Interspecific Hybrids Between and. G3 (2020) doi:10.1534/g3.120.401427.